The key role and challenges of cell apoptosis in cancer treatment
Apoptosis is a strictly regulated and evolutionarily conserved cell death program, also known for its role as a tumor suppressive mechanism. Apoptosis is a normal physiological cell death response to many stimuli, infections, or injuries, including those caused by cytotoxic drug therapy or radiation therapy regimens, which can result in irreparable DNA damage. The role of apoptosis in cancer has received widespread attention, and resistance to apoptosis is widely recognized as an acquired characteristic of cancer cells, giving them survival advantages, promoting tumor evolution and growth, and leading to treatment failure. Therefore, the efficacy of cancer treatment not only depends strictly on the cell damage they cause, but also on the ability of cells to activate their apoptosis program. In recent decades, research on cancer treatment has mainly focused on developing improved drugs and radiation therapy, aiming to induce maximum tumor cell death, thereby reducing tumor volume and blocking invasiveness.
1. Apoptosis and breast cancer
The clustering of death receptors (DRs) at the membrane leads to apoptosis. Björn Högberg, Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Sweden, and his team present a stimuli-responsive robotic switch nanodevice that can autonomously and selectively turn on the display of cytotoxic ligand patterns in tumour microenvironments[1]. They demonstrate a switchable DNA origami that normally hides six ligands but displays them as a hexagonal pattern 10 nm in diameter once under higher acidity (Fig.1). This can effectively cluster DRs and trigger apoptosis of human breast cancer cells at pH 6.5 while remaining inert at pH 7.4. When administered to mice bearing human breast cancer xenografts, this nanodevice decreased tumour growth by up to 70%. The data demonstrate the feasibility and opportunities for developing ligand pattern switches as a path for targeted treatment.
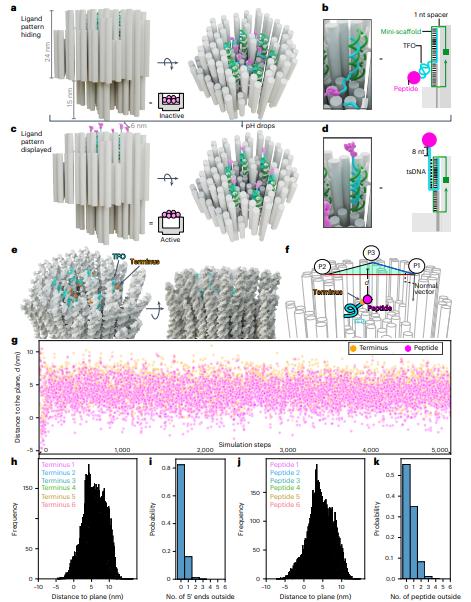
Fig.1 Design principles and simulations of the pH-responsive origami switch for ligand pattern hiding and display
2. Apoptosis and Colorectal cancer
Resistance to immunotherapy in colorectal cancer (CRC) is associated with obstruction of FAS (Apo‐1 or CD95)‐dependent apoptosis. Qinxi Li, Faculty of Medicine and Life Sciences, Xiamen University, China, and his team demonstrated that the upregulation of pirin (PIR) protein in colon cancers promotes tumorigenesis[2]. Knockout or inhibition of PIR dramatically increases FAS expression, FAS-dependent apoptosis and attenuates colorectal tumor formation in mice. Specifically, NFκB2 is a direct transcriptional activator of FAS and robustly suppressed by PIR in dual mechanisms. One is the disruption of NFκB2 complex (p52-RELB) association with FAS promoter, the other is the inhibition of NIK-mediated NFκB2 activation and nuclear translocation, leading to the inability of active NFκB2 complex toward the transcription of FAS (Fig.2). Furthermore, PIR interacts with FAS and recruits it in cytosol, preventing its membrane translocation and assembling. Importantly, knockdown or knockout of PIR dramatically sensitizes cells to FAS mAb- or active CD8+ T cells triggered cell death. Taken together, a PIR-NIK-NFκB2-FAS survival pathway is established, which plays a key role in supporting CRC survival.
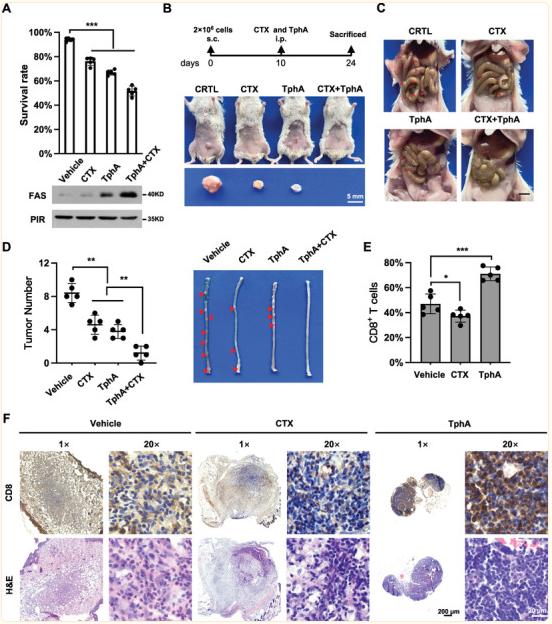
Fig.2 PIR suppresses FAS-dependent apoptosis by switching off NFκB2-FAS axis
3. Apoptosis and bladder cancer
Molecular understanding of muscle-invasive (MIBC) and non–muscle-invasive (NMIBC) bladder cancer is currently based primarily on transcriptomic and genomic analyses. François Radvanyi, Institut Curie, PSL Research University, Paris, France, and his team obtained proteomic data from 40 MIBC and 23 NMIBC cases, and proteomic groups from unsupervised analyses (uPGs) were characterized using clinicopathological, proteomic, genomic, transcriptomic, and pathway enrichment analyses[3]. Five uPGs, covering both NMIBC and MIBC, were identified and uPG-E was associated with the Ta pathway and enriched in FGFR3 mutations. Their analyses also highlighted enrichment of proteins involved in apoptosis in FGFR3-mutated tumors. Genetic and pharmacological inhibition demonstrated that FGFR3 activation regulates TRAIL receptor expression and sensitizes cells to TRAIL-mediated apoptosis, further increased by combination with birinapant (Fig.3). This proteogenomic study provides a comprehensive resource for investigating NMIBC and MIBC heterogeneity and highlights the potential of TRAIL-induced apoptosis as a treatment option for FGFR3-mutated bladder tumors, warranting a clinical investigation.
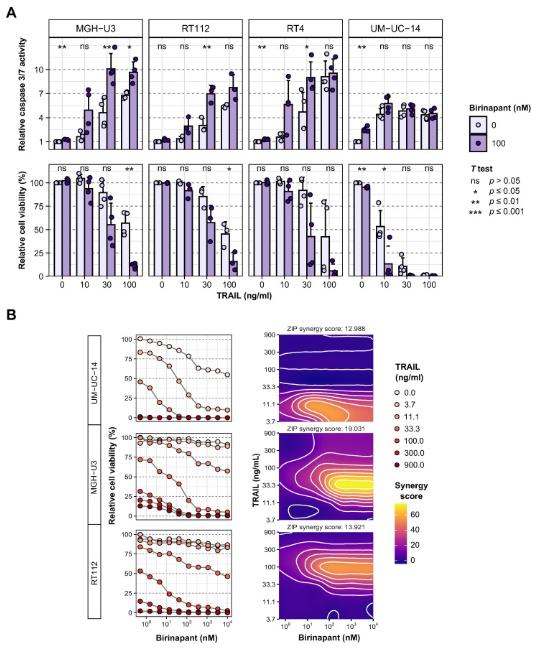
Fig.3 Combination of TRAIL with apoptotic sensitizer birinapant is synergistic in FGFR3-altered urothelial cell lines
4. Apoptosis and blood cancer
TP53-mutant blood cancers remain a clinical challenge. BH3-mimetic drugs inhibit BCL-2 pro-survival proteins, inducing cancer cell apoptosis. Despite acting downstream of p53, functional p53 is required for maximal cancer cell killing by BH3-mimetics through an unknown mechanism. Gemma L. Kelly, The Walter and Eliza Hall Institute of Medical Research, Australia, and his team reported p53 is activated following BH3-mimetic induced mitochondrial outer membrane permeabilization, leading to BH3-only protein induction and thereby potentiating the pro-apoptotic signal[4]. TP53-deficient lymphomas lack this feedforward loop, providing opportunities for survival and disease relapse after BH3-mimetic treatment. The therapeutic barrier imposed by defects in TP53 can be overcome by direct activation of the cGAS/STING pathway, which promotes apoptosis of blood cancer cells through p53-independent BH3-only protein upregulation (Fig.4). Combining clinically relevant STING agonists with BH3-mimetic drugs efficiently kills TRP53/TP53-mutant mouse B lymphoma, human NK/T lymphoma, and acute myeloid leukemia cells. This represents a promising therapy regime that can be fast-tracked to tackle TP53-mutant blood cancers in the clinic.
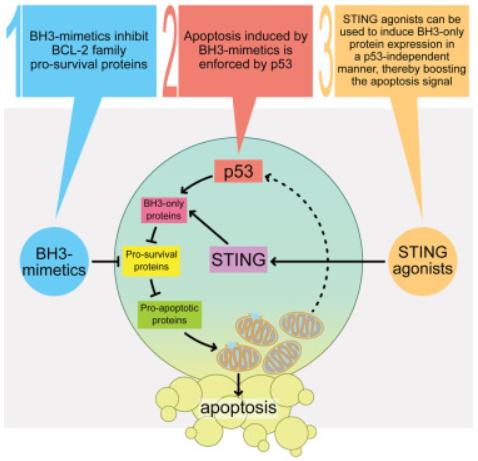
Fig.4 STING pathway activation triggers p53-independent apoptosis in blood cancers
Cloud-Clone can not only provide a variety of tumor experimental animal models, including tumor transplant animal models, spontaneous tumor animal models, induced tumor animal models, tumor metastasis animal models, etc., covering common tumor research. We also have various cancer detection, cell apoptosis related products, and the above DR, FAS, pirin, NF κ B2, FGFR3, TRAIL, p53, STING and other related products, which can help the scientific researchers to carry out research on cell apoptosis and cancer.
References
[1]Wang Y, Baars I, Berzina I, et al. A DNA robotic switch with regulated autonomous display of cytotoxic ligand nanopatterns. Nat Nanotechnol. 2024. doi:10.1038/s41565-024-01676-4. (IF=38.1)
[2]Ma H, Suleman M, Zhang F, et al. Pirin Inhibits FAS-Mediated Apoptosis to Support Colorectal Cancer Survival. Adv Sci (Weinh). 2024;11(10):e2301476. (IF=14.3)
[3]Groeneveld CS, Sanchez-Quiles V, Dufour F, et al. Proteogenomic Characterization of Bladder Cancer Reveals Sensitivity to Apoptosis Induced by Tumor Necrosis Factor-related Apoptosis-inducing Ligand in FGFR3-mutated Tumors. Eur Urol. 2024;85(5):483-494. (IF=25.3)
[4]Diepstraten ST, Yuan Y, La Marca JE, et al. Putting the STING back into BH3-mimetic drugs for TP53-mutant blood cancers. Cancer Cell. 2024;42(5):850-868.e9. (IF=48.8)
