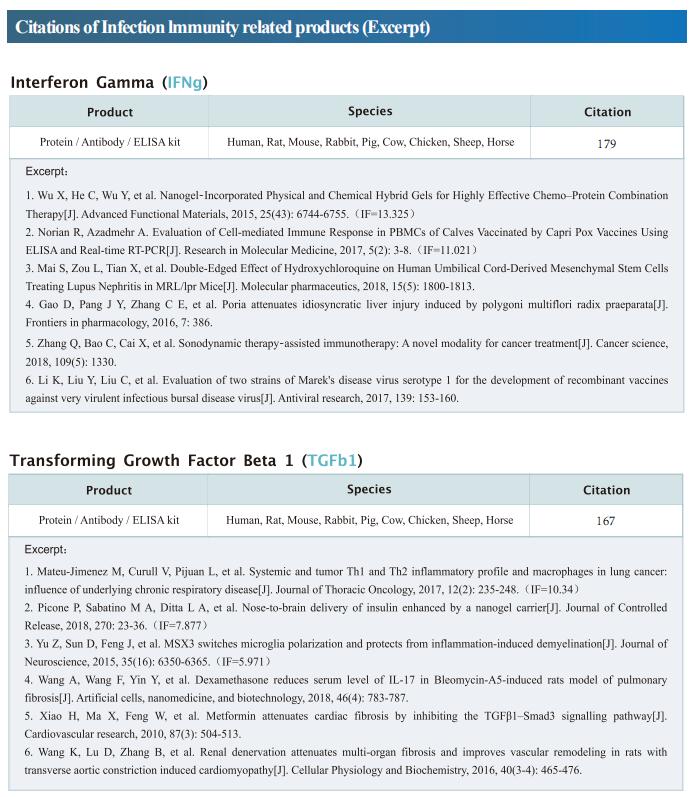
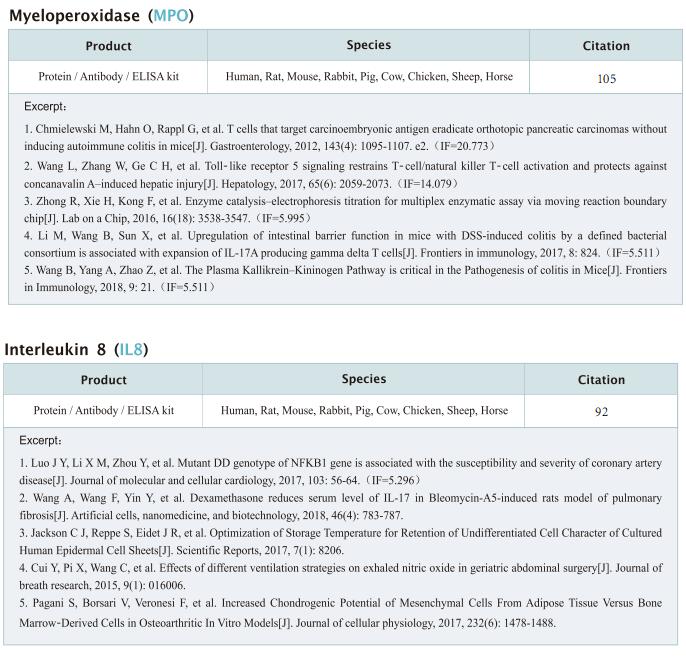
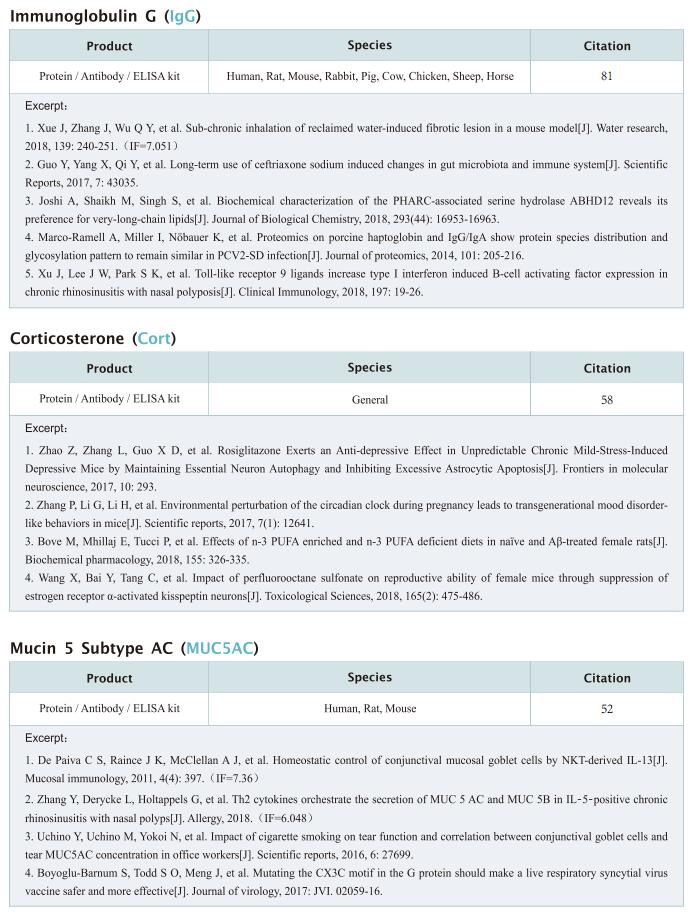
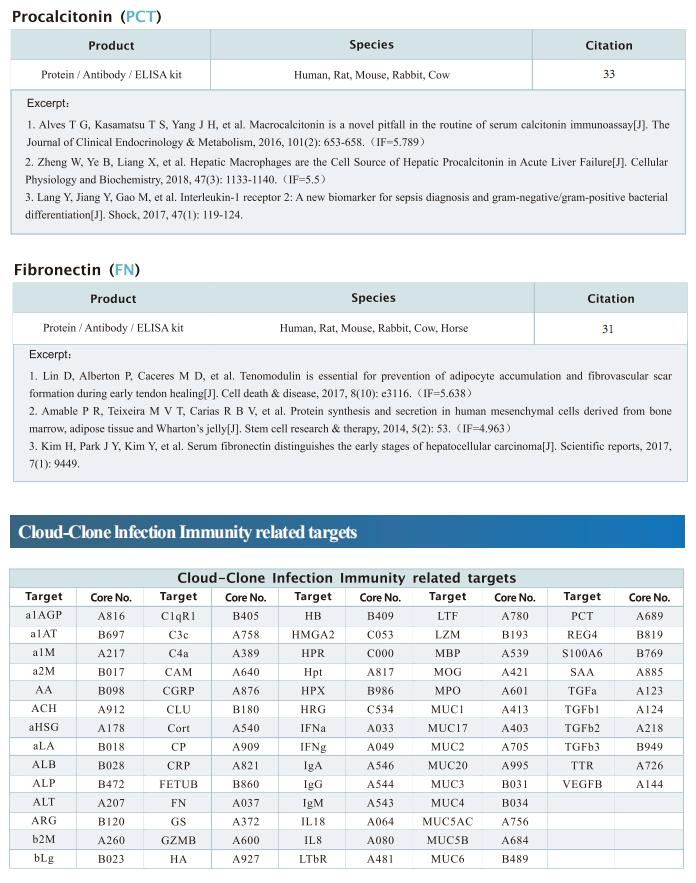
New progress in HIV research
Acquired immunodeficiency syndrome (AIDS) is an infectious disease caused by human immunodeficiency virus (HIV) infection, which seriously endangers people’s health and lives. At present, the clinical treatment for AIDS is antiretroviral therapy (ART), which minimizes viral replication in patients and reduces plasma viral load to a level that cannot be detected by conventional testing methods. However, once the treatment is stopped, viral load will rebound to the pre-treatment level. An important reason why HIV is difficult to eliminate from the body is the long-term existence of HIV latent reservoir. The HIV latent virus reservoir composed of latently infected cells is a huge obstacle that cannot be eliminated by current clinical treatments. Therefore, it is necessary to conduct further research on the molecular mechanisms underlying HIV-1 latency.
1. FBXO34 promotes latent HIV-1 activation by posttranscriptional modulation
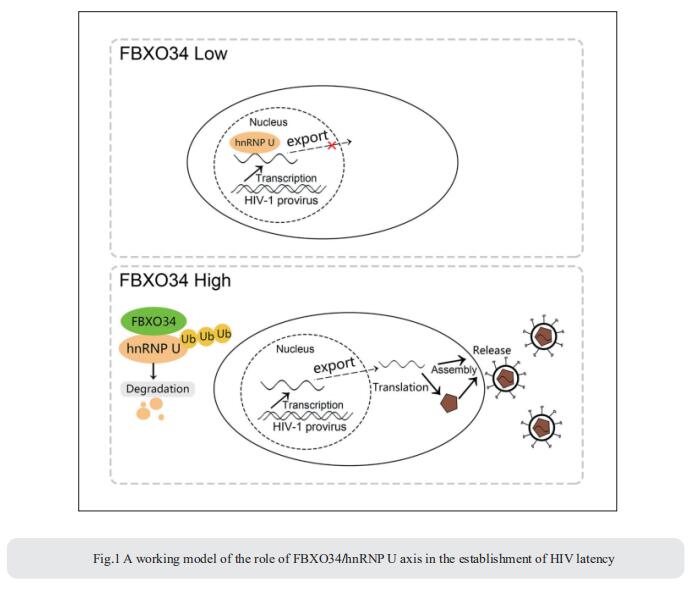
To eliminate latent virus in infected cells, it is necessary to have a clear understanding of the mechanism of HIV-1 latency and maintenance. Huanzhang Zhu, Institute of Genetics, School of Life Sciences, Fudan University, China, and his team identified E3 ubiquitin ligase F-box protein 34 (FBXO34) and the substrate of FBXO34, heterogeneous nuclear ribonucleoprotein U (hnRNP U) was identified by affinity purification mass spectrometry, as new host factors related to HIV-1 latent maintenance[1]. Overexpression of FBXO34 or knockout of hnRNP U can activate latent HIV-1 in multiple latent cell lines. FBXO34 mainly promotes hnRNP U ubiquitination, which leads to hnRNP U degradation and abolishment of the interaction between hnRNP U and HIV-1 mRNA. In a latently infected cell line, hnRNP U interacts with the ReV region of HIV-1 mRNA through amino acids 1-339 to hinder HIV-1 translation, thereby, promoting HIV-1 latency (Fig.1). They confirmed the role of the FBXO34/hnRNP U axis in the primary CD4+ T lymphocyte model, and detected differences in hnRNP U expression levels in samples from patients treated with antiretroviral therapy (ART) and healthy people, which further suggests that the FBXO34/hnRNP U axis is a new pathway involved in HIV-1 latency. This novel FBXO34/hnRNP U axis in HIV transcription may be directly targeted to control HIV reservoirs in patients in the future.
2. Epigenetic silencing by the SMC5/6 complex mediates HIV-1 latency
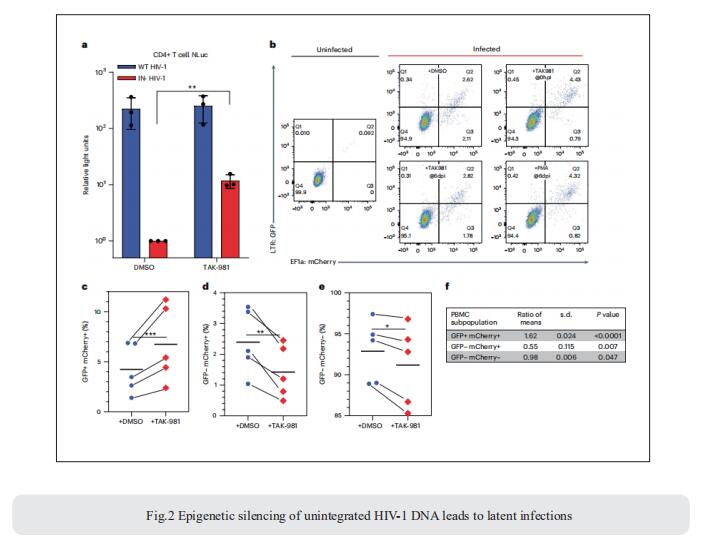
After viral entry and reverse transcription, HIV-1 proviruses that fail to integrate are epigenetically silenced, but the underlying mechanism has remained unclear. Bryan R. Cullen, Department of Molecular Genetics and Microbiology, Duke University Medical Center, USA, and his team identifed the host SMC5/6 complex as essential for this epigenetic silencing by using a genome-wide CRISPR/Cas9 knockout screen[2]. They showed that SMC5/6 binds to and then SUMOylates unintegrated chromatinized HIV-1 DNA. Inhibition of SUMOylation, either by point mutagenesis of the SMC5/6 component NSMCE2—a SUMO E3 ligase—or using the SUMOylation inhibitor TAK-981, prevents epigenetic silencing, enables transcription from unintegrated HIV-1 DNA and rescues the replication of integrase-defcient HIV-1. Finally, they showed that blocking SMC5/6 complex expression, or inhibiting its SUMOylation activity, suppresses the establishment of latent HIV-1 infections in both CD4+ T cell lines and primary human T cells (Fig.2). The data show that the SMC5/6 complex plays a direct role in mediating the establishment of HIV-1 latency by epigenetically silencing integration-competent HIV-1 proviruses before integration.
3. Single-cell multiomics reveals persistence of HIV-1 in expanded cytotoxic T cell clones
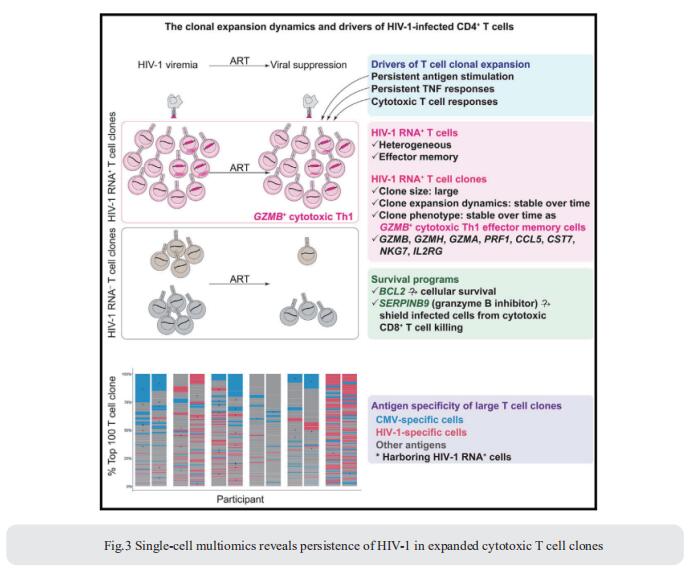
Understanding the drivers and markers of clonally expanding HIV-1-infected CD4+ T cells is essential for HIV-1 eradication. Ya-Chi Ho, Department of Microbial Pathogenesis, Yale University School of Medicine, USA, and his team used single-cell ECCITE-seq, which captures surface protein expression, cellular transcriptome, HIV-1 RNA, and TCR sequences within the same single cell to track clonal expansion dynamics in longitudinally archived samples from six HIV-1-infected individuals (during viremia and after suppressive antiretroviral therapy) and two uninfected individuals[3]. Despite antiretroviral therapy, persistent antigen and TNF responses shaped T cell clonal expansion. HIV-1 resided in Th1-polarized, antigen-responding T cells expressing BCL2 and SERPINB9 that may resist cell death. HIV-1 RNA+ T cell clones were larger in clone size, established during viremia, persistent after viral suppression, and enriched in GZMB+ cytotoxic effector memory Th1 cells (Fig.3). Targeting HIV-1-infected cytotoxic CD4+ T cells and drivers of clonal expansion provides another direction for HIV-1 eradication.
References
[1]Yang X, Zhao X, Zhu Y, et al. FBXO34 promotes latent HIV-1 activation by post-transcriptional modulation [J]. Emerg Microbes Infect. 2022;11(1):2785-2799. (IF=19.568)
[2]Irwan ID, Bogerd HP, Cullen BR. Epigenetic silencing by the SMC5/6 complex mediates HIV-1 latency [J]. Nat Microbiol. 2022;7(12):2101-2113. (IF=30.964)
[3]Collora JA, Liu R, Pinto-Santini D, et al. Single-cell multiomics reveals persistence of HIV-1 in expanded cytotoxic T cell clones [J]. Immunity. 2022;55(6):1013-1031.e7. (IF=43.474)
Cloud-Clone not only provides various products related to commonly used infection immune detection indicators, including PCT, CRP, CGRP, IL-6, IL-8, IL-10, HMGB1, SAA, IFNα, etc. We also have HIV-1 recombinant protein and the above FBXO34/hnRNP U, BCL2, SERPINB9, GZMB and other related protein detection products, which can help the majority of scientific researchers to carry out HIV-1 infection related research.